| PolarSPARC |
Introduction to Apache Parquet
| Bhaskar S | 10/07/2022 |
Overview
Data collected over a period of time (in any domain and referred to as a data set) is often used to perform data analysis for gaining insights and for future predictions. One can store the data set in CSV files for future consumption.
Data analysis typically focuses on a limited set of columns from the rows of a data set. This implies reading all the rows from the CSV file before pruning unwanted columns from the data set. What if the data set contains millions (or billions) of rows ???
Is there a better and efficient way to store large data sets for future data processing ???
Please welcome the honorable storage format - the Apache Parquet !!!
Apache Parquet is an open-source data storage format that uses an hybrid of row-column oriented storage for efficient storage and retrieval of data AND work with any choice of data processing framework, data model, or programming language.
Given a data set (of rows and columns), there are two common data processing patterns:
Online Transaction Processing (or OLTP for short) is a type of data processing, where a large number of business transactions (inserts, updates, deletes) on different rows of the data set, are executing concurrently in realtime
Online Analytical Processing (or OLAP for short) is a type of data processing, where data aggregated from various OLTP sources is stored centrally to query and perform analysis on the data from different points of view (roll-up, slicing, dicing, pivoting)
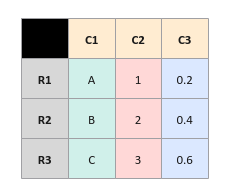
To get a better understanding, let us consider the following sample data set of rows (R1, R2, R3) and columns (C1, C2, C3):

Since OLTP processing works on a row at a time (inserts, updates, deletes), it is more efficient to store the data set in a row oriented format as shown in the illustration below:

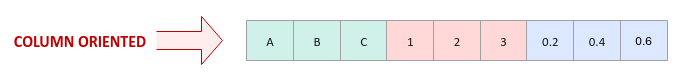
On the other hand, OLAP processing works by performing selections (queries with criteria) on few columns from the rows and hence, it is more efficient to store the data set in a column oriented format as shown in the illustration below:
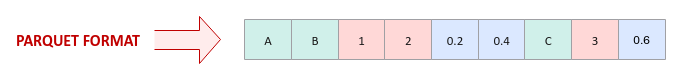
The Parquet format is a hybrid of the row oriented and the column oriented storage, where columns for a chunk of rows (with metadata) is stored together as shown in the illustration below:
The hybrid storage format not only enables for efficient data storage but also for efficient data retrieval using selection criteria.
Parquet Format Internals
In order to better understand the Parquet file format, we will use the storage layout as shown in the illustration below:
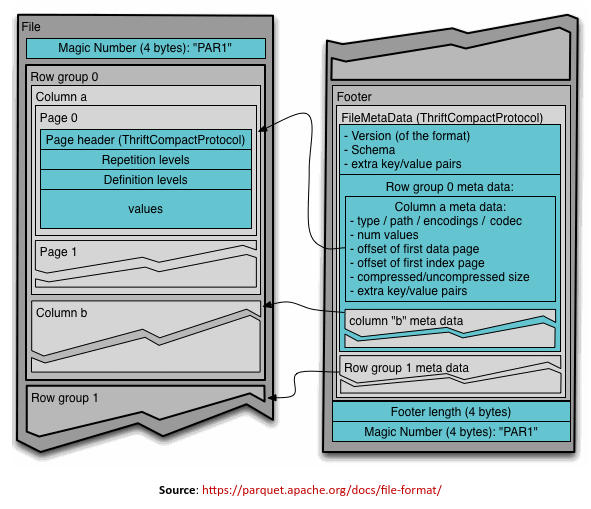
A Parquet file starts with a file header, followed by one or more Row Groups, and ending with a file footer (referred to as the FileMetaData). A Row Group in turn consists of a data chunk for every column in the row of the data set (often referred to as the Column Chunk). The Column Chunk in turn is made up of one or more data Pages which contains the actual column data along with some metadata.
The Parquet file format described above is visually shown in the illustration below:
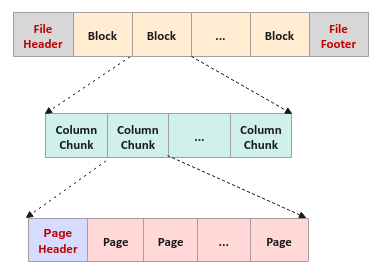
The following are some of the core parts of the file storage format:
Page is an indivisible chunk of data values for a specific column from the rows of the data set with a specific size (default 1 MB). In addition, every Page consists of a page header that includes some metadata information such as the number of data values in the page, the maximum and the minimum value of the column data values in the page, etc. Given that a Page only contains data for a single column of the data set, one can enable data compression such as the Dictionary Encoding or Run Length Encoding
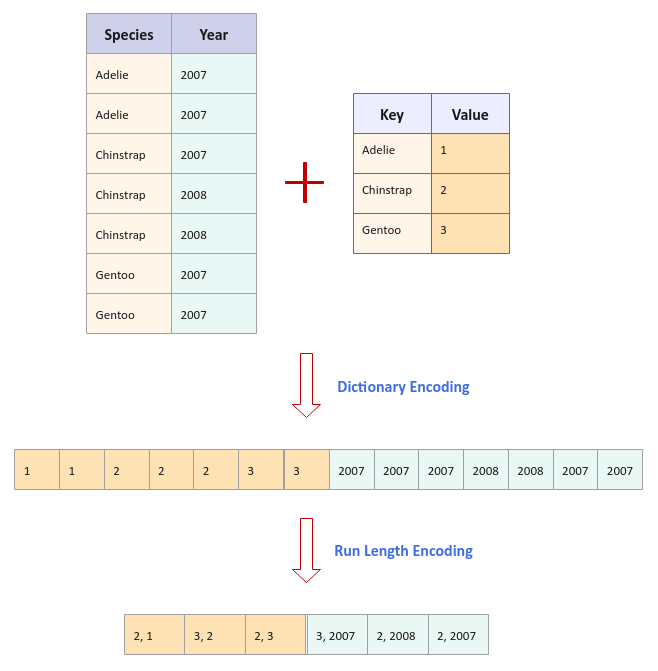
With Dictionary Encoding, repeating text values can be encoded using integer values to conserve space. For example, if we take the three species of Penguins - Adelie, Chinstrap, and Gentoo, they can be assigned integer values of 1, 2, and 3 respectively. So, instead of storing the text for the Penguin species, we can store their associated integer values. This approach results in significant storage space.
With Run Length Encoding, a sequence of repeating numbers can be replaced by the count of their occurence followed by the actual number value. For example, if we had the sequence 2007, 2007, 2007, 2008, 2008, 2007, and 2007. They could be encoded as (3, 2007), (2, 2008), and (2, 2007) respectively, where the encoding format is of the form (count, value).
The following visual depicts the Dictionary and Run Length Encoding with an example:
Column Chunk represents a chunk of data for a particular column from the rows of the data set
Row Group is a logical partitioning of the rows of a data set into a block of specific size (default of 128MB). It consists of a Column Chunk for every column from the rows of the data set
FileMetaData includes metadata information such as the schema of the data set, the various Column Chunks and their starting offsets in the file.
The following illustration shows the structure of the FileMetaData:
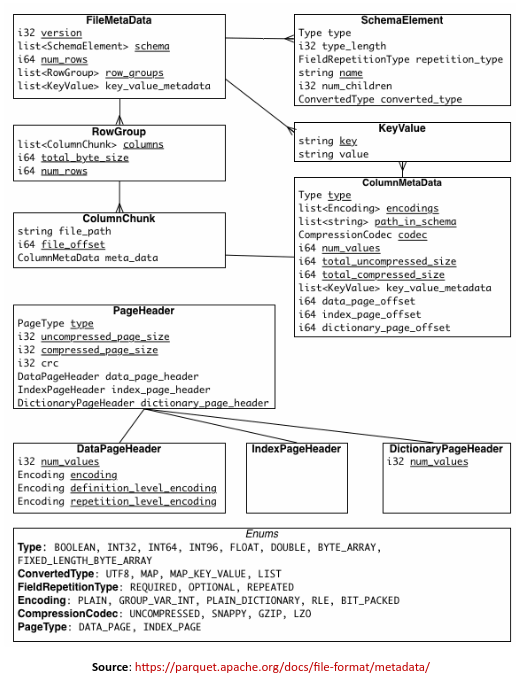
The metadata in the parquet file allows for fast traversal (with minimal I/O) to the desired columns (using FileMetaData) and the within the columns to filter out values that do not match the criteria (using the Page statistics).
Installation and Setup
The installation is on a Ubuntu 22.04 LTS based Linux desktop.
We need to install the following python packages - pandas, pyarrow, and parquet-cli from the Ubuntu repository.
To install the mentioned packages, execute the following commands:
$ sudo apt update
$ sudo apt install pandas pyarrow parquet-cli -y
The package parquet-cli provides a basic command-line tool called parq for exploring parquet files.
Hands-on with Parquet
For the demonstration, we will create a parquet file from the Palmer Penguins data set using pandas. The following is the Python code to create three types of file formats - csv, uncompressed parquet, and compressed parquet files:
#
# @Author: Bhaskar S
# @Blog: https://www.polarsparc.com
# @Date: 07 Oct 2022
#
import logging
import pandas as pd
logging.basicConfig(format='%(levelname)s %(asctime)s - %(message)s', level=logging.INFO)
# Load the Palmer Penguins data set, clean missing values and return a dataframe to the data set
def get_palmer_penguins():
logging.info('Ready to load the Palmer Penguins dataset...')
url = 'https://vincentarelbundock.github.io/Rdatasets/csv/palmerpenguins/penguins.csv'
df = pd.read_csv(url)
logging.info('Ready to cleanse the Palmer Penguins dataset...')
df = df.drop(df.columns[0], axis=1)
df = df.drop([3, 271], axis=0)
df.loc[[8, 10, 11], 'sex'] = 'female'
df.at[9, 'sex'] = 'male'
df.at[47, 'sex'] = 'female'
df.loc[[178, 218, 256, 268], 'sex'] = 'female'
logging.info('Completed cleansing the Palmer Penguins dataset...')
return df
# Create an uncompressed csv file
def write_uncompressed_csv(path, df):
logging.info('Ready to write the Palmer Penguins dataset as csv...')
df.to_csv(path, compression=None, index=False)
logging.info('Completed writing the Palmer Penguins dataset as csv...')
# Create an uncompressed parquet file
def write_uncompressed_parquet(path, df):
logging.info('Ready to write the Palmer Penguins dataset as Uncompressed Parquet...')
df.to_parquet(path, compression=None, index=False)
logging.info('Completed writing the Palmer Penguins dataset as Uncompressed Parquet...')
# Create a compressed parquet file
def write_compressed_parquet(path, df):
logging.info('Ready to write the Palmer Penguins dataset as Compressed Parquet...')
df.to_parquet(path, compression='snappy', index=False)
logging.info('Completed writing the Palmer Penguins dataset as Compressed Parquet...')
def main():
penguins_df = get_palmer_penguins()
write_uncompressed_csv('./data/unc_pp.csv', penguins_df)
write_uncompressed_parquet('./data/unc_pp.parquet', penguins_df)
write_compressed_parquet('./data/com_pp.parquet', penguins_df)
if __name__ == '__main__':
main()
Executing the above Python program sample-1.py will generate the three files in the subdirectory data. The following output shows the listing of the three files:
-rw-rw-r-- 1 polarsparc polarsparc 8761 Oct 7 19:37 com_pp.parquet -rw-rw-r-- 1 polarsparc polarsparc 16736 Oct 7 19:37 unc_pp.csv -rw-rw-r-- 1 polarsparc polarsparc 10234 Oct 7 19:37 unc_pp.parquet
Notice the file sizes of the three files. The compressed parquet file is the most compact.
We will now examine the compressed parquet file using the command-line tool parq.
To display the metadata information, execute the following command:
$ parq ./data/com_pp.parquet
The following would be the typical output:
# Metadata <pyarrow._parquet.FileMetaData object at 0x7f5fdc6de660> created_by: parquet-cpp-arrow version 9.0.0 num_columns: 8 num_rows: 342 num_row_groups: 1 format_version: 2.6 serialized_size: 4302
To display the schema information, execute the following command:
$ parq ./data/com_pp.parquet --schema
The following would be the typical output:
# Schema
<pyarrow._parquet.ParquetSchema object at 0x7f9748cc2cc0>
required group field_id=-1 schema {
optional binary field_id=-1 species (String);
optional binary field_id=-1 island (String);
optional double field_id=-1 bill_length_mm;
optional double field_id=-1 bill_depth_mm;
optional double field_id=-1 flipper_length_mm;
optional double field_id=-1 body_mass_g;
optional binary field_id=-1 sex (String);
optional int64 field_id=-1 year;
}
To display the first 5 rows from the data set, execute the following command:
$ parq ./data/com_pp.parquet --head 5
The following would be the typical output:
species island bill_length_mm bill_depth_mm flipper_length_mm \
0 Adelie Torgersen 39.1 18.7 181.0
1 Adelie Torgersen 39.5 17.4 186.0
2 Adelie Torgersen 40.3 18.0 195.0
3 Adelie Torgersen 36.7 19.3 193.0
4 Adelie Torgersen 39.3 20.6 190.0
body_mass_g sex year
0 3750.0 male 2007
1 3800.0 female 2007
2 3250.0 female 2007
3 3450.0 female 2007
4 3650.0 male 2007
To display the last 5 rows from the data set, execute the following command:
$ parq ./data/com_pp.parquet --tail 5
The following would be the typical output:
species island bill_length_mm bill_depth_mm flipper_length_mm \
337 Chinstrap Dream 55.8 19.8 207.0
338 Chinstrap Dream 43.5 18.1 202.0
339 Chinstrap Dream 49.6 18.2 193.0
340 Chinstrap Dream 50.8 19.0 210.0
341 Chinstrap Dream 50.2 18.7 198.0
body_mass_g sex year
337 4000.0 male 2009
338 3400.0 female 2009
339 3775.0 male 2009
340 4100.0 male 2009
341 3775.0 female 2009
The following is the Python code to programmatically display information (using pyarrow) about the two parquet files (uncompressed and compressed):
#
# @Author: Bhaskar S
# @Blog: https://www.polarsparc.com
# @Date: 07 Oct 2022
#
import logging
import pyarrow.parquet as pq
logging.basicConfig(format='%(levelname)s %(asctime)s - %(message)s', level=logging.INFO)
def read_uncompressed_parquet(path):
logging.info('Ready to read the Palmer Penguins dataset from Uncompressed Parquet...\n')
pq_unc = pq.ParquetFile(path)
logging.info('Schema -> %s', pq_unc.schema)
logging.info('No. of Row Groups -> %d\n', pq_unc.num_row_groups)
logging.info('Metadata -> %s\n', pq_unc.metadata)
logging.info('First Row Group (Metadata) -> %s\n', pq_unc.metadata.row_group(0))
logging.info('First Row Group (Content) -> %s\n', pq_unc.read_row_group(0))
logging.info('First Row Group, Second Column Chunk (Metadata) -> %s', pq_unc.metadata.row_group(0).column(1))
logging.info('Completed reading the Palmer Penguins dataset from Uncompressed Parquet...')
def read_compressed_parquet(path):
logging.info('Ready to read the Palmer Penguins dataset from Compressed Parquet...\n')
pq_com = pq.ParquetFile(path)
logging.info('Schema -> %s', pq_com.schema)
logging.info('No. of Row Groups -> %d\n', pq_com.num_row_groups)
logging.info('Metadata -> %s\n', pq_com.metadata)
logging.info('First Row Group (Metadata) -> %s\n', pq_com.metadata.row_group(0))
logging.info('First Row Group (Content) -> %s\n', pq_com.read_row_group(0))
logging.info('First Row Group, Second Column Chunk (Metadata) -> %s', pq_com.metadata.row_group(0).column(1))
logging.info('Completed reading the Palmer Penguins dataset from Compressed Parquet...')
def main():
read_uncompressed_parquet('./data/unc_pp.parquet')
logging.info('\n')
logging.info('--------------------------------------------------\n')
read_compressed_parquet('./data/com_pp.parquet')
if __name__ == '__main__':
main()
Executing the above Python program sample-2.py will produce the following output:
INFO 2022-10-07 19:39:23,799 - Ready to read the Palmer Penguins dataset from Uncompressed Parquet...
INFO 2022-10-07 19:39:23,800 - Schema -> <pyarrow._parquet.ParquetSchema object at 0x7f2d64991e80>
required group field_id=-1 schema {
optional binary field_id=-1 species (String);
optional binary field_id=-1 island (String);
optional double field_id=-1 bill_length_mm;
optional double field_id=-1 bill_depth_mm;
optional double field_id=-1 flipper_length_mm;
optional double field_id=-1 body_mass_g;
optional binary field_id=-1 sex (String);
optional int64 field_id=-1 year;
}
INFO 2022-10-07 19:39:23,800 - No. of Row Groups -> 1
INFO 2022-10-07 19:39:23,800 - Metadata -> <pyarrow._parquet.FileMetaData object at 0x7f2d60181210>
created_by: parquet-cpp-arrow version 9.0.0
num_columns: 8
num_rows: 342
num_row_groups: 1
format_version: 2.6
serialized_size: 4302
INFO 2022-10-07 19:39:23,800 - First Row Group (Metadata) -> <pyarrow._parquet.RowGroupMetaData object at 0x7f2d5bf46bb0>
num_columns: 8
num_rows: 342
total_byte_size: 5174
INFO 2022-10-07 19:39:23,824 - First Row Group (Content) -> pyarrow.Table
species: string
island: string
bill_length_mm: double
bill_depth_mm: double
flipper_length_mm: double
body_mass_g: double
sex: string
year: int64
----
species: [["Adelie","Adelie","Adelie","Adelie","Adelie",...,"Chinstrap","Chinstrap","Chinstrap","Chinstrap","Chinstrap"]]
island: [["Torgersen","Torgersen","Torgersen","Torgersen","Torgersen",...,"Dream","Dream","Dream","Dream","Dream"]]
bill_length_mm: [[39.1,39.5,40.3,36.7,39.3,...,55.8,43.5,49.6,50.8,50.2]]
bill_depth_mm: [[18.7,17.4,18,19.3,20.6,...,19.8,18.1,18.2,19,18.7]]
flipper_length_mm: [[181,186,195,193,190,...,207,202,193,210,198]]
body_mass_g: [[3750,3800,3250,3450,3650,...,4000,3400,3775,4100,3775]]
sex: [["male","female","female","female","male",...,"male","female","male","male","female"]]
year: [[2007,2007,2007,2007,2007,...,2009,2009,2009,2009,2009]]
INFO 2022-10-07 19:39:23,824 - First Row Group, Second Column Chunk (Metadata) -> <pyarrow._parquet.ColumnChunkMetaData object at 0x7f2d601d72e0>
file_offset: 299
file_path:
physical_type: BYTE_ARRAY
num_values: 342
path_in_schema: island
is_stats_set: True
statistics:
<pyarrow._parquet.Statistics object at 0x7f2d5bf6a750>
has_min_max: True
min: Biscoe
max: Torgersen
null_count: 0
distinct_count: 0
num_values: 342
physical_type: BYTE_ARRAY
logical_type: String
converted_type (legacy): UTF8
compression: UNCOMPRESSED
encodings: ('RLE_DICTIONARY', 'PLAIN', 'RLE')
has_dictionary_page: True
dictionary_page_offset: 180
data_page_offset: 226
total_compressed_size: 119
total_uncompressed_size: 119
INFO 2022-10-07 19:39:23,824 - Completed reading the Palmer Penguins dataset from Uncompressed Parquet...
INFO 2022-10-07 19:39:23,824 -
INFO 2022-10-07 19:39:23,824 - --------------------------------------------------
INFO 2022-10-07 19:39:23,825 - Ready to read the Palmer Penguins dataset from Compressed Parquet...
INFO 2022-10-07 19:39:23,825 - Schema -> <pyarrow._parquet.ParquetSchema object at 0x7f2d64e9c0c0>
required group field_id=-1 schema {
optional binary field_id=-1 species (String);
optional binary field_id=-1 island (String);
optional double field_id=-1 bill_length_mm;
optional double field_id=-1 bill_depth_mm;
optional double field_id=-1 flipper_length_mm;
optional double field_id=-1 body_mass_g;
optional binary field_id=-1 sex (String);
optional int64 field_id=-1 year;
}
INFO 2022-10-07 19:39:23,825 - No. of Row Groups -> 1
INFO 2022-10-07 19:39:23,825 - Metadata -> <pyarrow._parquet.FileMetaData object at 0x7f2d601d74c0>
created_by: parquet-cpp-arrow version 9.0.0
num_columns: 8
num_rows: 342
num_row_groups: 1
format_version: 2.6
serialized_size: 4302
INFO 2022-10-07 19:39:23,825 - First Row Group (Metadata) -> <pyarrow._parquet.RowGroupMetaData object at 0x7f2d5bf46bb0>
num_columns: 8
num_rows: 342
total_byte_size: 5174
INFO 2022-10-07 19:39:23,825 - First Row Group (Content) -> pyarrow.Table
species: string
island: string
bill_length_mm: double
bill_depth_mm: double
flipper_length_mm: double
body_mass_g: double
sex: string
year: int64
----
species: [["Adelie","Adelie","Adelie","Adelie","Adelie",...,"Chinstrap","Chinstrap","Chinstrap","Chinstrap","Chinstrap"]]
island: [["Torgersen","Torgersen","Torgersen","Torgersen","Torgersen",...,"Dream","Dream","Dream","Dream","Dream"]]
bill_length_mm: [[39.1,39.5,40.3,36.7,39.3,...,55.8,43.5,49.6,50.8,50.2]]
bill_depth_mm: [[18.7,17.4,18,19.3,20.6,...,19.8,18.1,18.2,19,18.7]]
flipper_length_mm: [[181,186,195,193,190,...,207,202,193,210,198]]
body_mass_g: [[3750,3800,3250,3450,3650,...,4000,3400,3775,4100,3775]]
sex: [["male","female","female","female","male",...,"male","female","male","male","female"]]
year: [[2007,2007,2007,2007,2007,...,2009,2009,2009,2009,2009]]
INFO 2022-10-07 19:39:23,826 - First Row Group, Second Column Chunk (Metadata) -> <pyarrow._parquet.ColumnChunkMetaData object at 0x7f2d5bf6b240>
file_offset: 306
file_path:
physical_type: BYTE_ARRAY
num_values: 342
path_in_schema: island
is_stats_set: True
statistics:
<pyarrow._parquet.Statistics object at 0x7f2d5bf6b290>
has_min_max: True
min: Biscoe
max: Torgersen
null_count: 0
distinct_count: 0
num_values: 342
physical_type: BYTE_ARRAY
logical_type: String
converted_type (legacy): UTF8
compression: SNAPPY
encodings: ('RLE_DICTIONARY', 'PLAIN', 'RLE')
has_dictionary_page: True
dictionary_page_offset: 183
data_page_offset: 231
total_compressed_size: 123
total_uncompressed_size: 119
INFO 2022-10-07 19:39:23,826 - Completed reading the Palmer Penguins dataset from Compressed Parquet...
The following is the Python code to programmatically (using pyarrow) select specific columns (species and body_mass_g) from the parquet file and filter rows using a criteria (body_mass_g > 4500):
#
# @Author: Bhaskar S
# @Blog: https://www.polarsparc.com
# @Date: 07 Oct 2022
#
import logging
import pyarrow.parquet as pq
logging.basicConfig(format='%(levelname)s %(asctime)s - %(message)s', level=logging.INFO)
def select_parquet_columns(file):
return pq.read_table(file, columns=['species', 'body_mass_g'])
def filter_parquet_columns(file):
return pq.read_table(file, columns=['species', 'body_mass_g'], filters=[('body_mass_g', '>', 4500)])
def main():
logging.info('Reading columns species and body_mass_g from the Palmer Penguins dataset from Parquet...')
df = select_parquet_columns('./data/com_pp.parquet').to_pandas()
logging.info('Displaying top 5 rows from the data set...\n')
logging.info(df.head(5))
logging.info('--------------------------------------------------\n')
logging.info('Filtering column body_mass_g > 4500 from the Palmer Penguins dataset from Parquet...')
df = filter_parquet_columns('./data/com_pp.parquet').to_pandas()
logging.info('Displaying top 5 rows from the filtered data set...\n')
logging.info(df.head(5))
if __name__ == '__main__':
main()
Executing the above Python program sample-3.py will produce the following output:
INFO 2022-10-07 19:41:28,135 - Reading columns species and body_mass_g from the Palmer Penguins dataset from Parquet... INFO 2022-10-07 19:41:28,344 - Displaying top 5 rows from the data set... INFO 2022-10-07 19:41:28,345 - species body_mass_g 0 Adelie 3750.0 1 Adelie 3800.0 2 Adelie 3250.0 3 Adelie 3450.0 4 Adelie 3650.0 INFO 2022-10-07 19:41:28,348 - -------------------------------------------------- INFO 2022-10-07 19:41:28,348 - Filtering column body_mass_g > 4500 from the Palmer Penguins dataset from Parquet... INFO 2022-10-07 19:41:28,351 - Displaying top 5 rows from the filtered data set... INFO 2022-10-07 19:41:28,351 - species body_mass_g 0 Adelie 4675.0 1 Adelie 4650.0 2 Adelie 4600.0 3 Adelie 4700.0 4 Adelie 4725.0
The following is the link to the Github Repo that provides the data files (csv, and parquet) as well as the Python code used in this article:
References